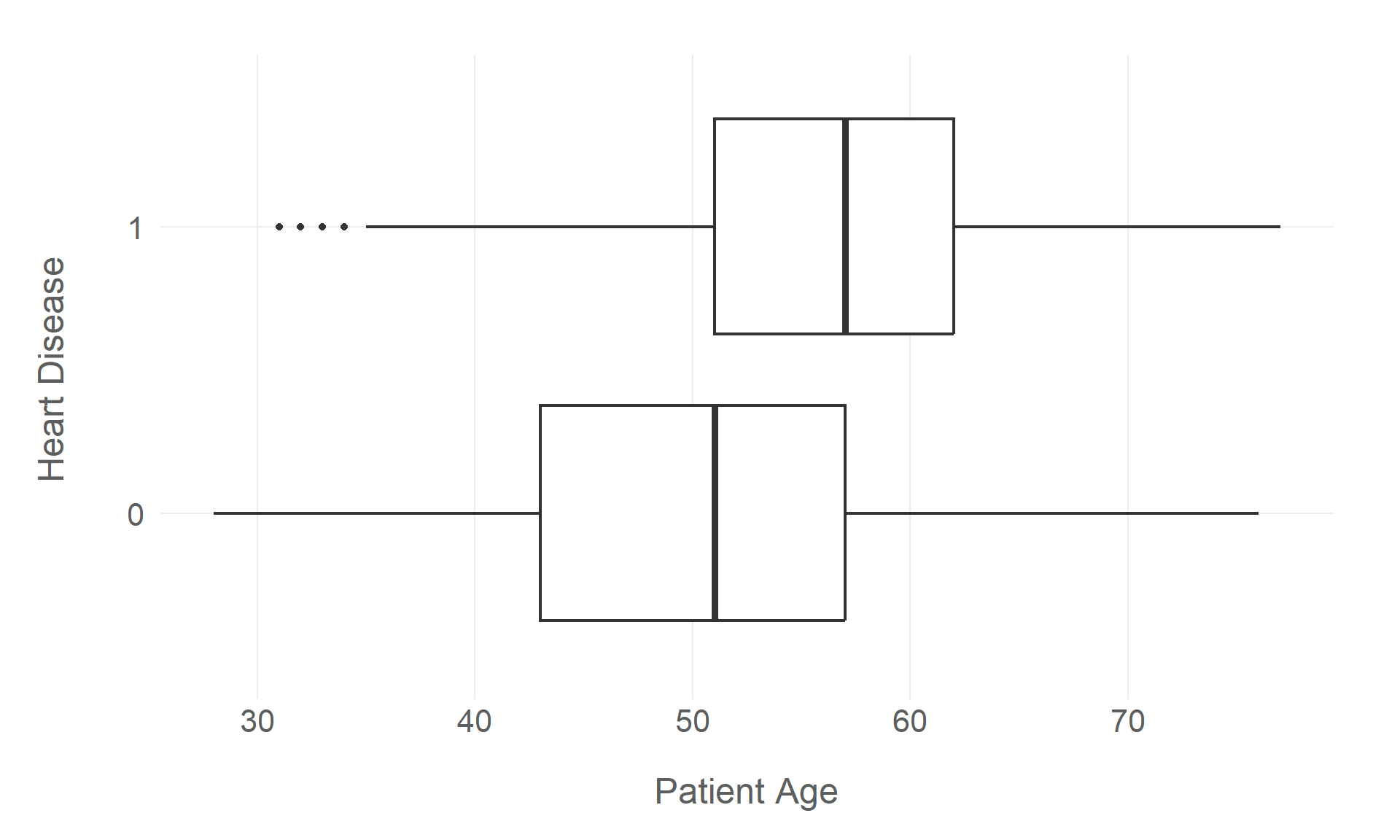
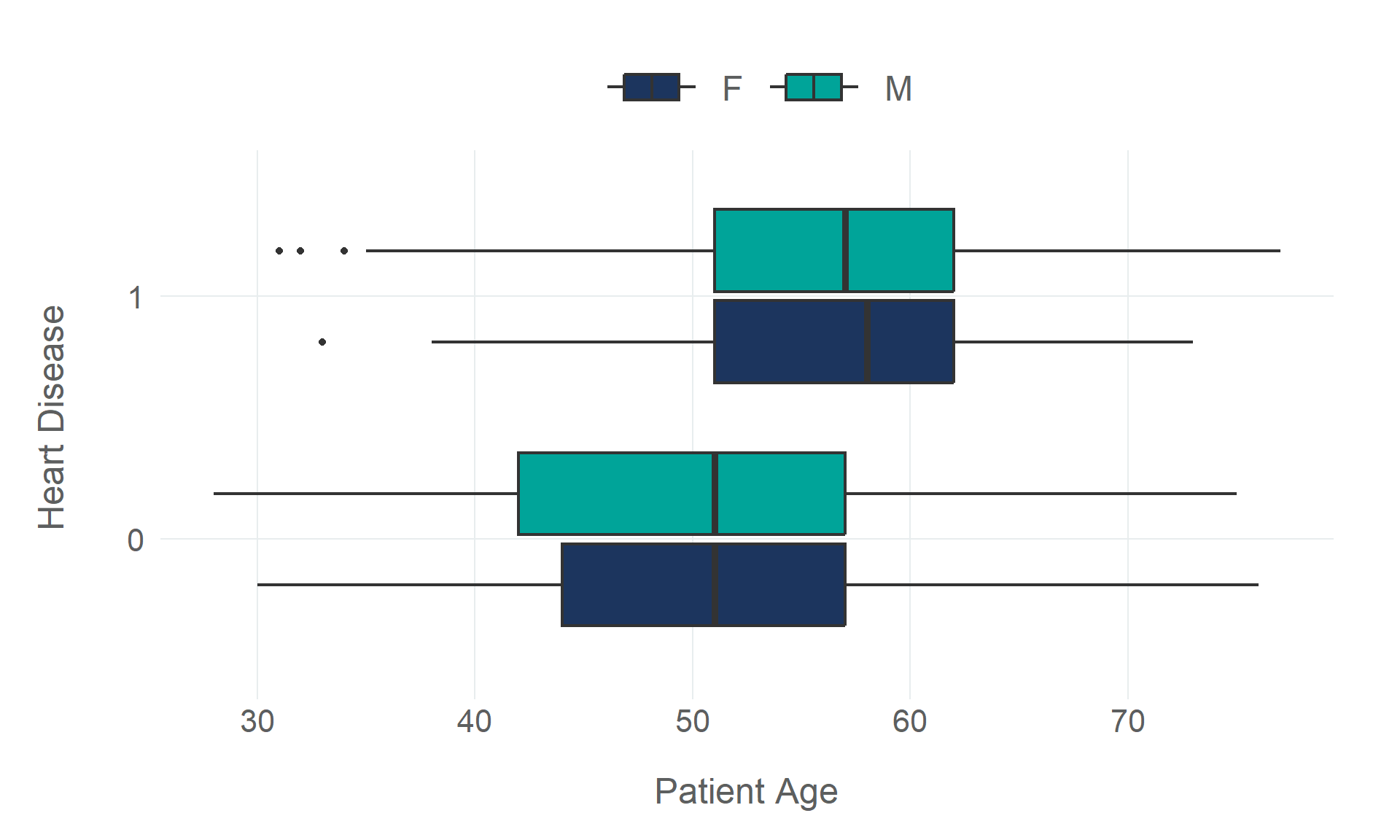
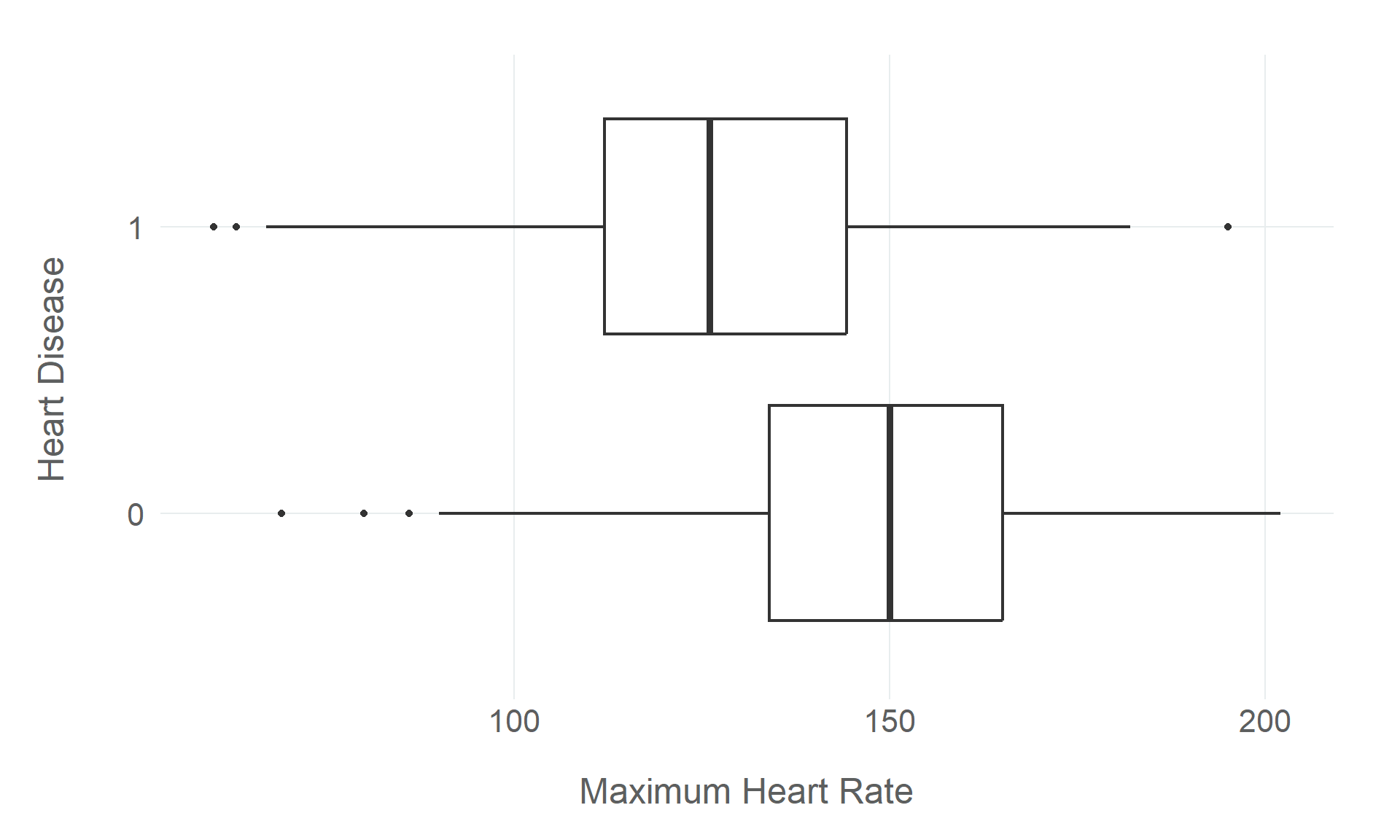
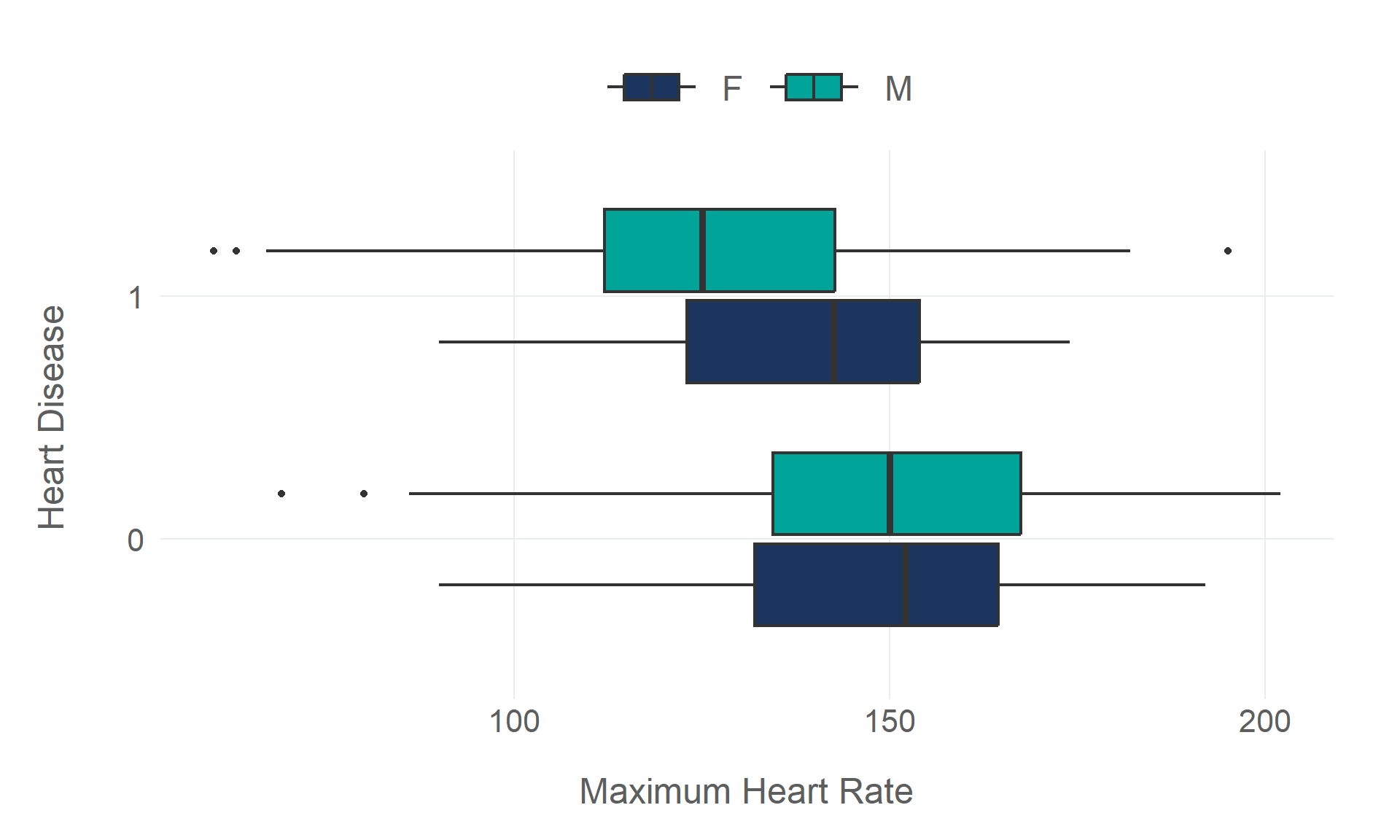
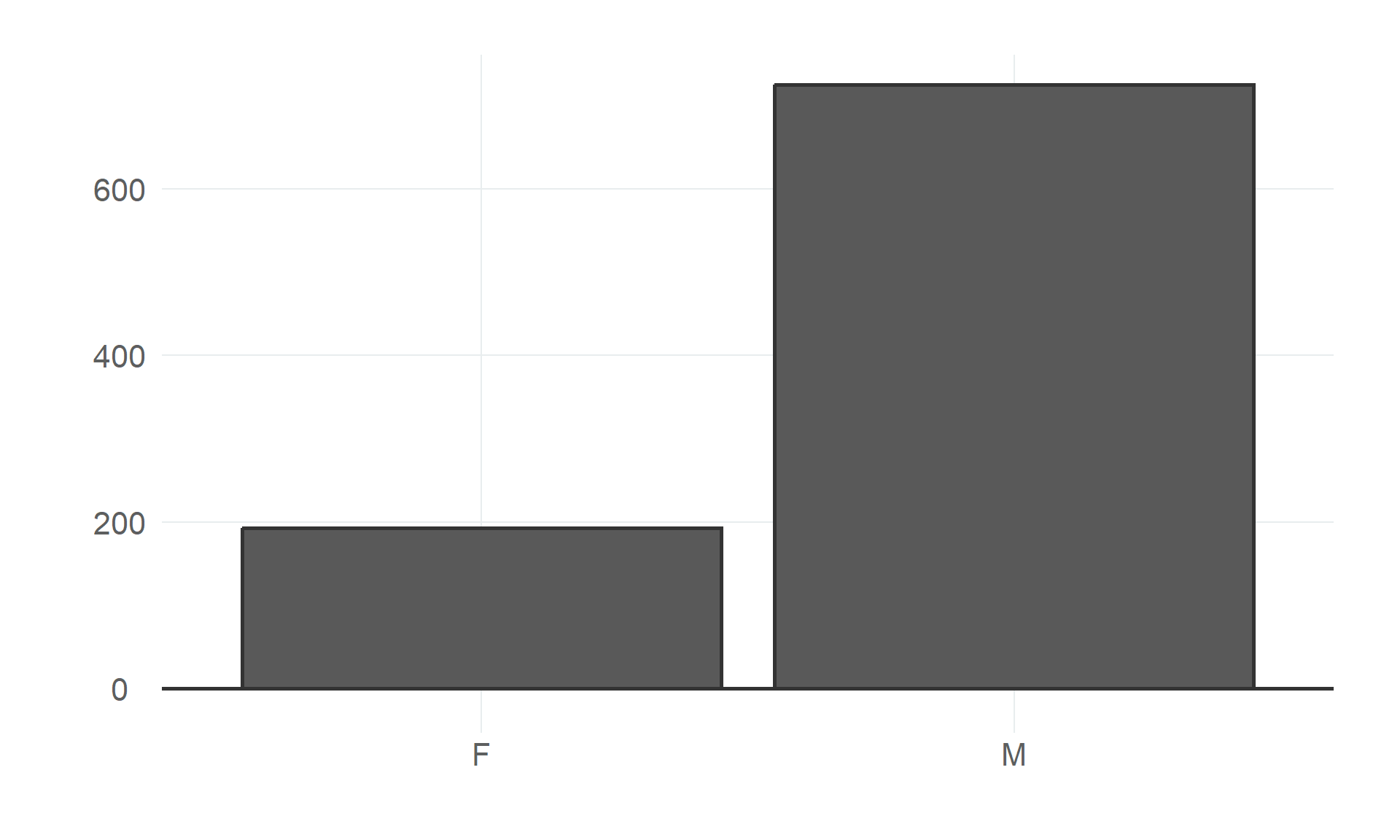Rows: 918
Columns: 10
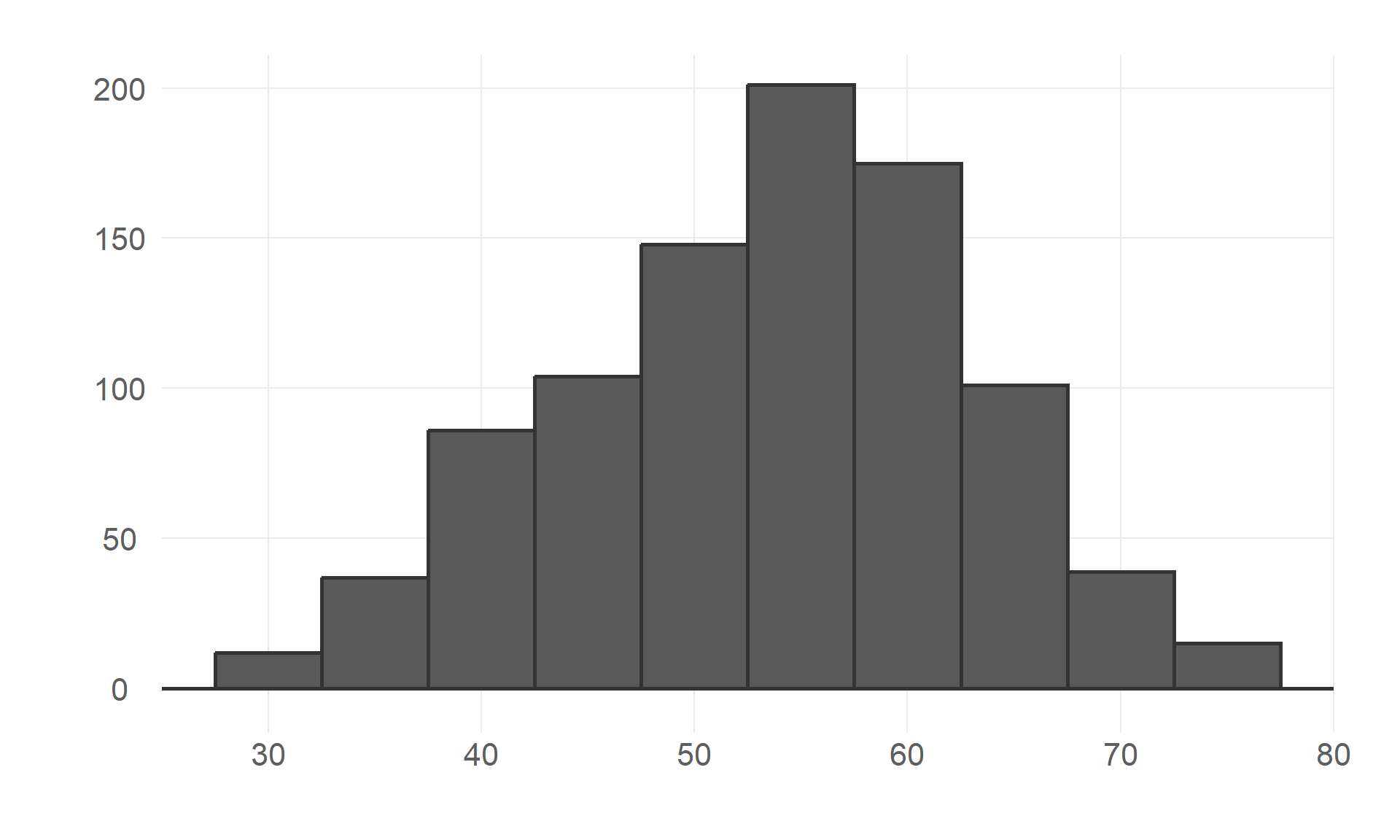
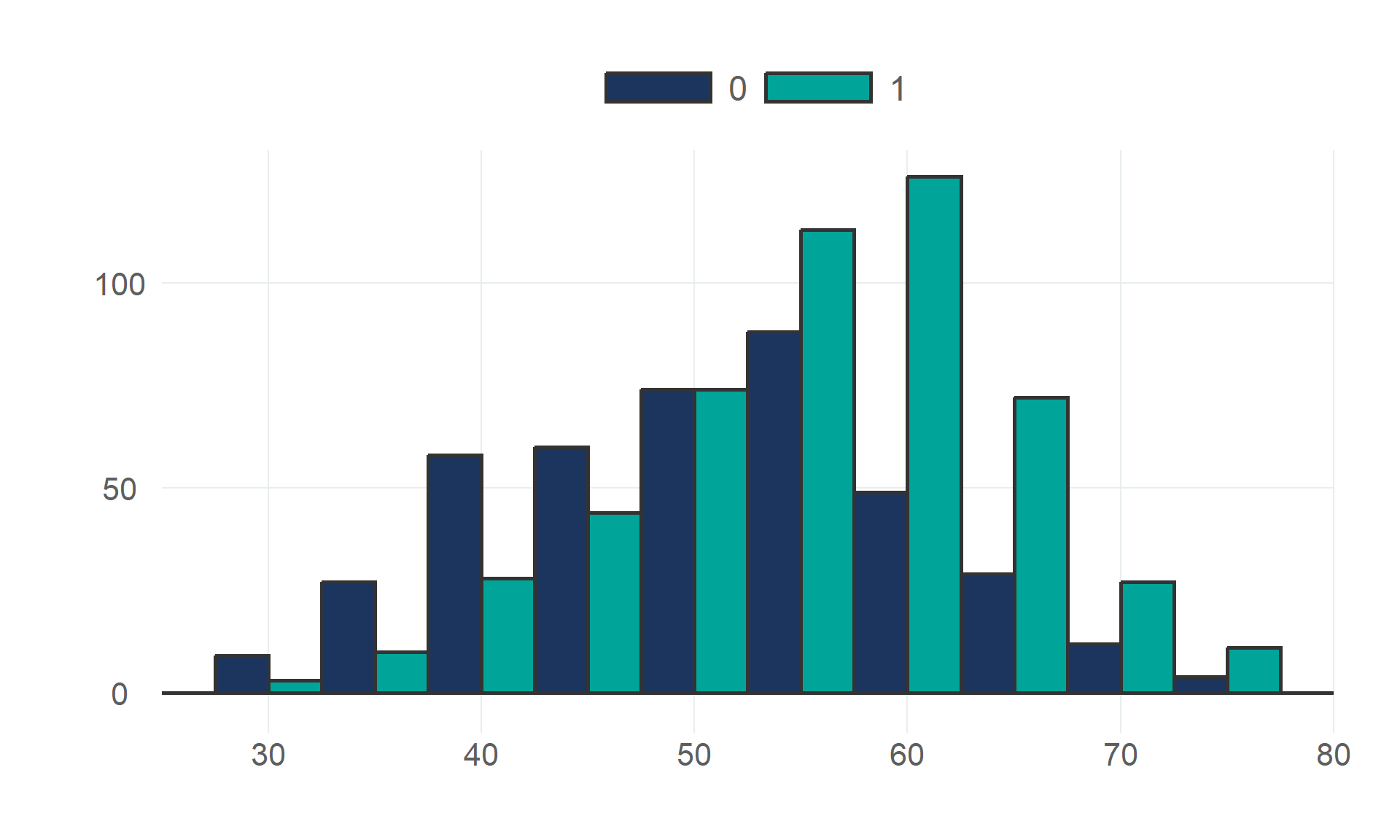
$ age <dbl> 40, 49, 37, 48, 54, 39, 45, 54, 37, 48, 37, 58, 39, 49, 42, 54, 38, 43, 60, 36, 43, 44, 49,…
$ sex <fct> M, F, M, F, M, M, F, M, M, F, F, M, M, M, F, F, M, F, M, M, F, M, F, M, M, M, M, M, F, M, M…
$ resting_bp <dbl> 140, 160, 130, 138, 150, 120, 130, 110, 140, 120, 130, 136, 120, 140, 115, 120, 110, 120, 1…
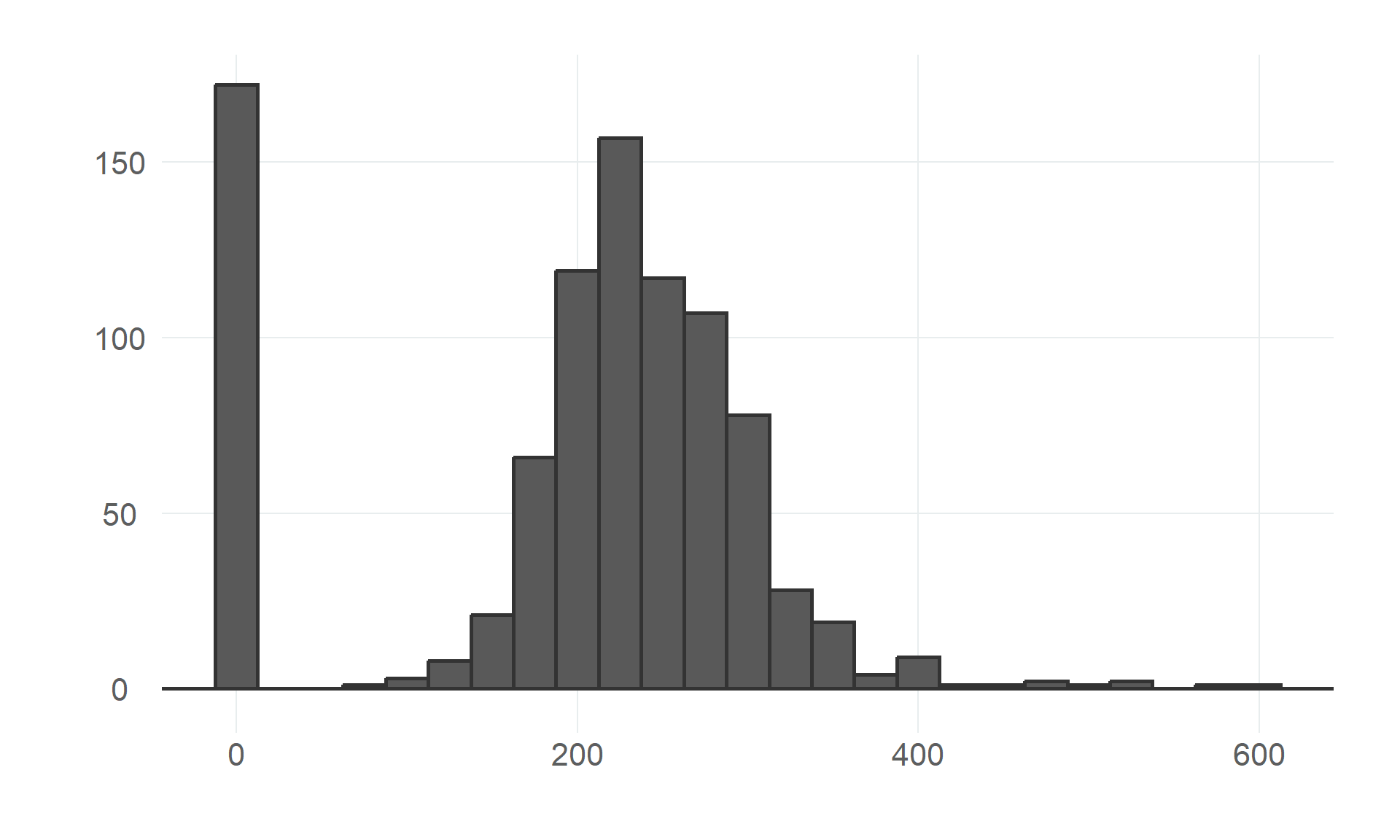
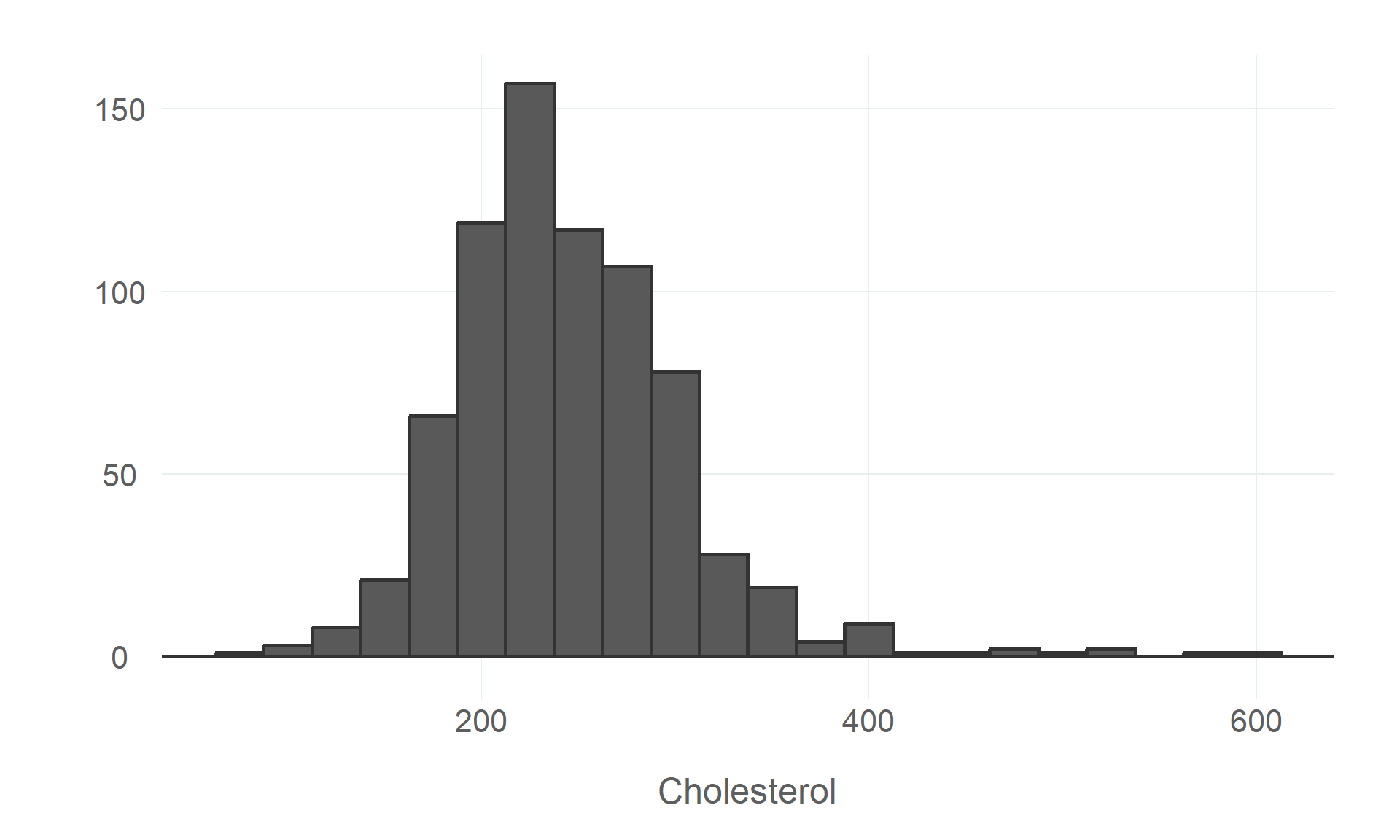
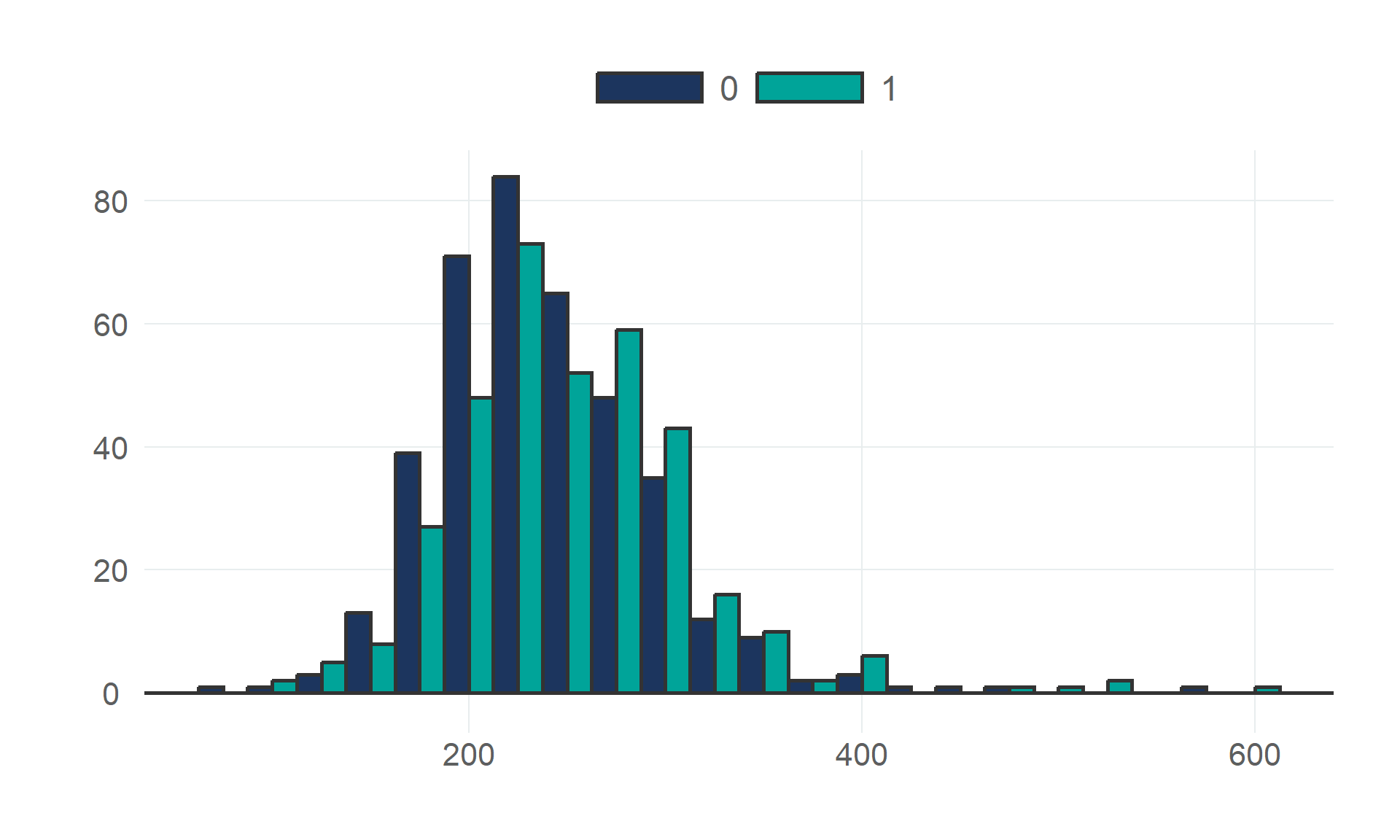
$ cholesterol <dbl> 289, 180, 283, 214, 195, 339, 237, 208, 207, 284, 211, 164, 204, 234, 211, 273, 196, 201, 2…
$ fasting_bs <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ resting_ecg <fct> Normal, Normal, ST, Normal, Normal, Normal, Normal, Normal, Normal, Normal, Normal, ST, Nor…
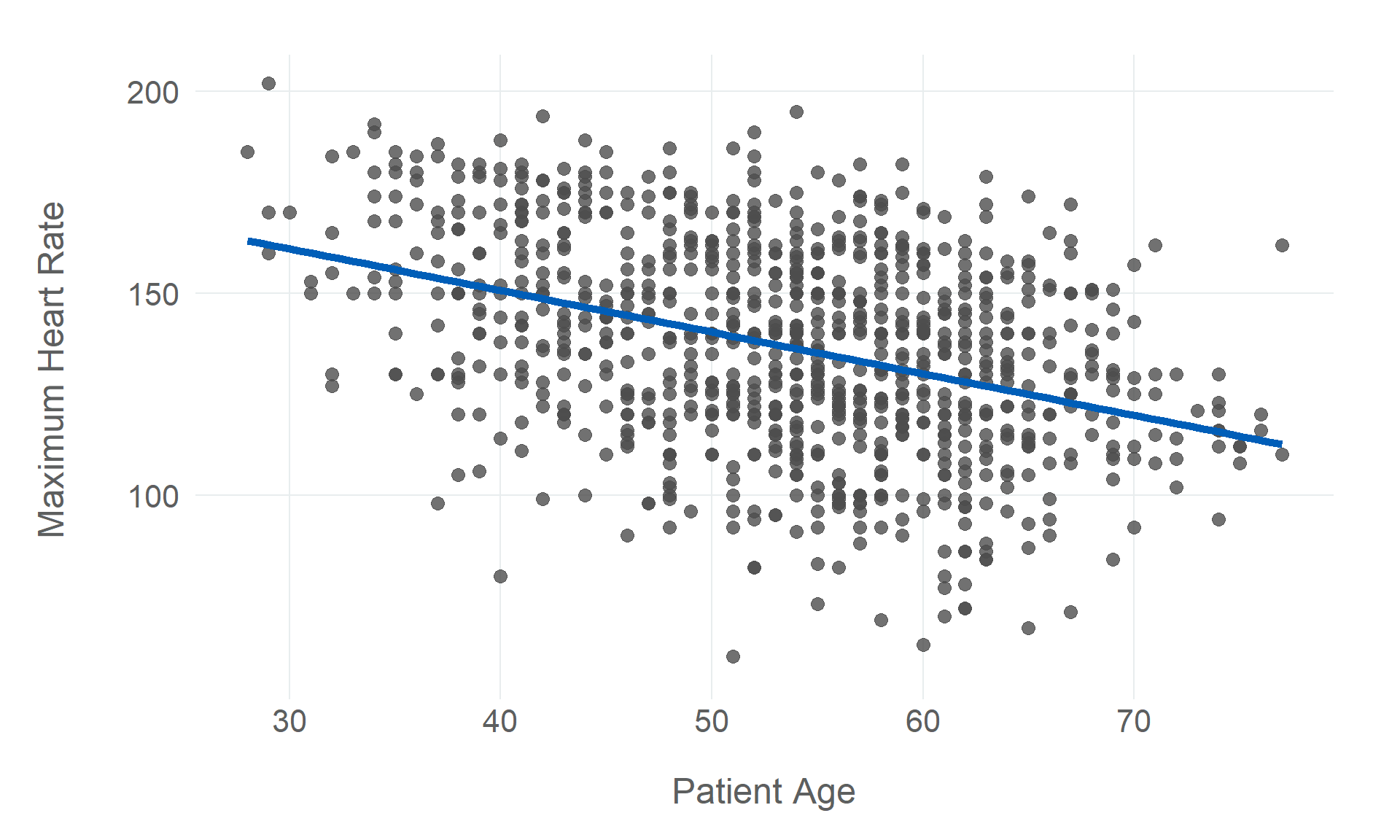
$ max_hr <dbl> 172, 156, 98, 108, 122, 170, 170, 142, 130, 120, 142, 99, 145, 140, 137, 150, 166, 165, 125…
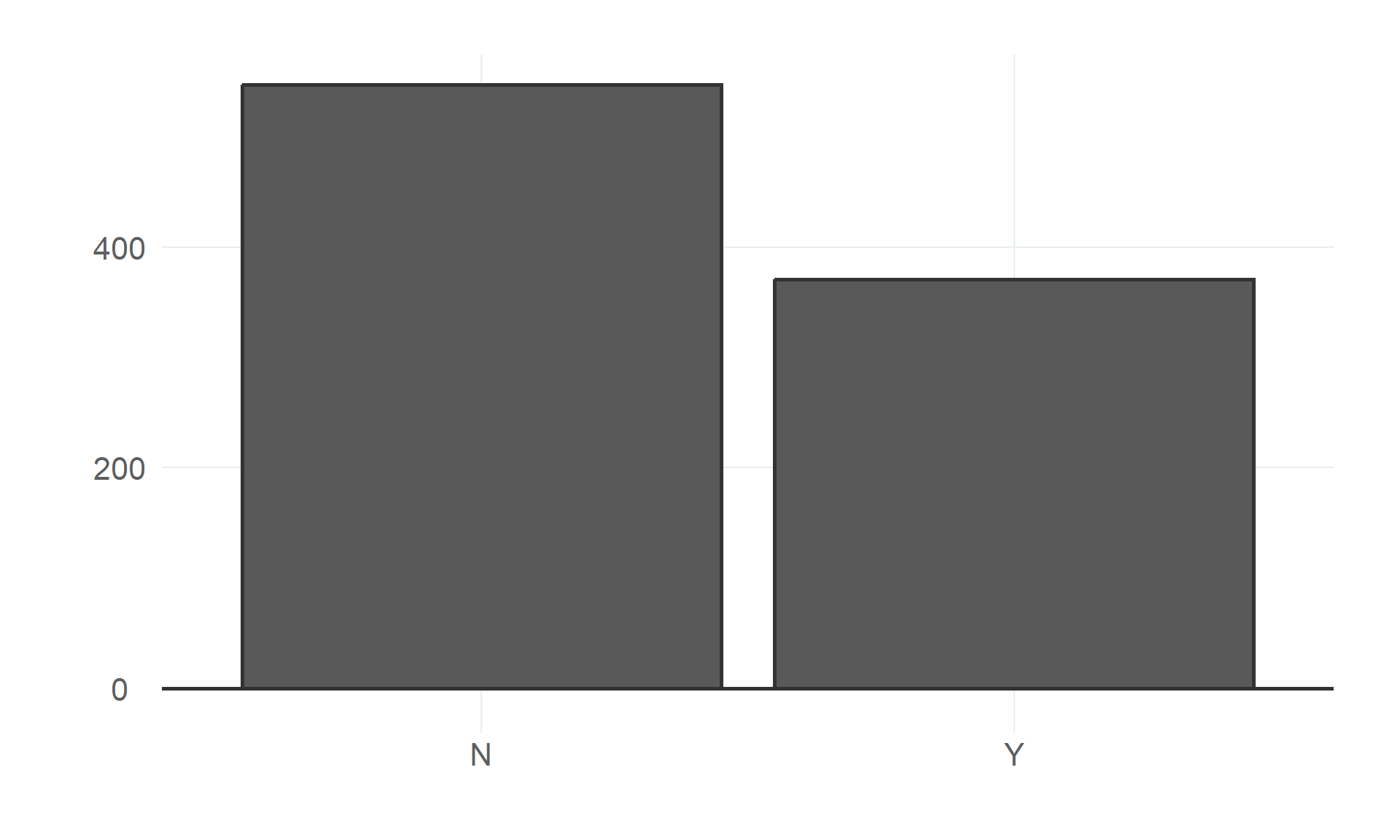
$ angina <fct> N, N, N, Y, N, N, N, N, Y, N, N, Y, N, Y, N, N, N, N, N, N, N, N, N, Y, N, N, Y, N, N, N, N…
$ heart_peak_reading <dbl> 0.0, 1.0, 0.0, 1.5, 0.0, 0.0, 0.0, 0.0, 1.5, 0.0, 0.0, 2.0, 0.0, 1.0, 0.0, 1.5, 0.0, 0.0, 1…
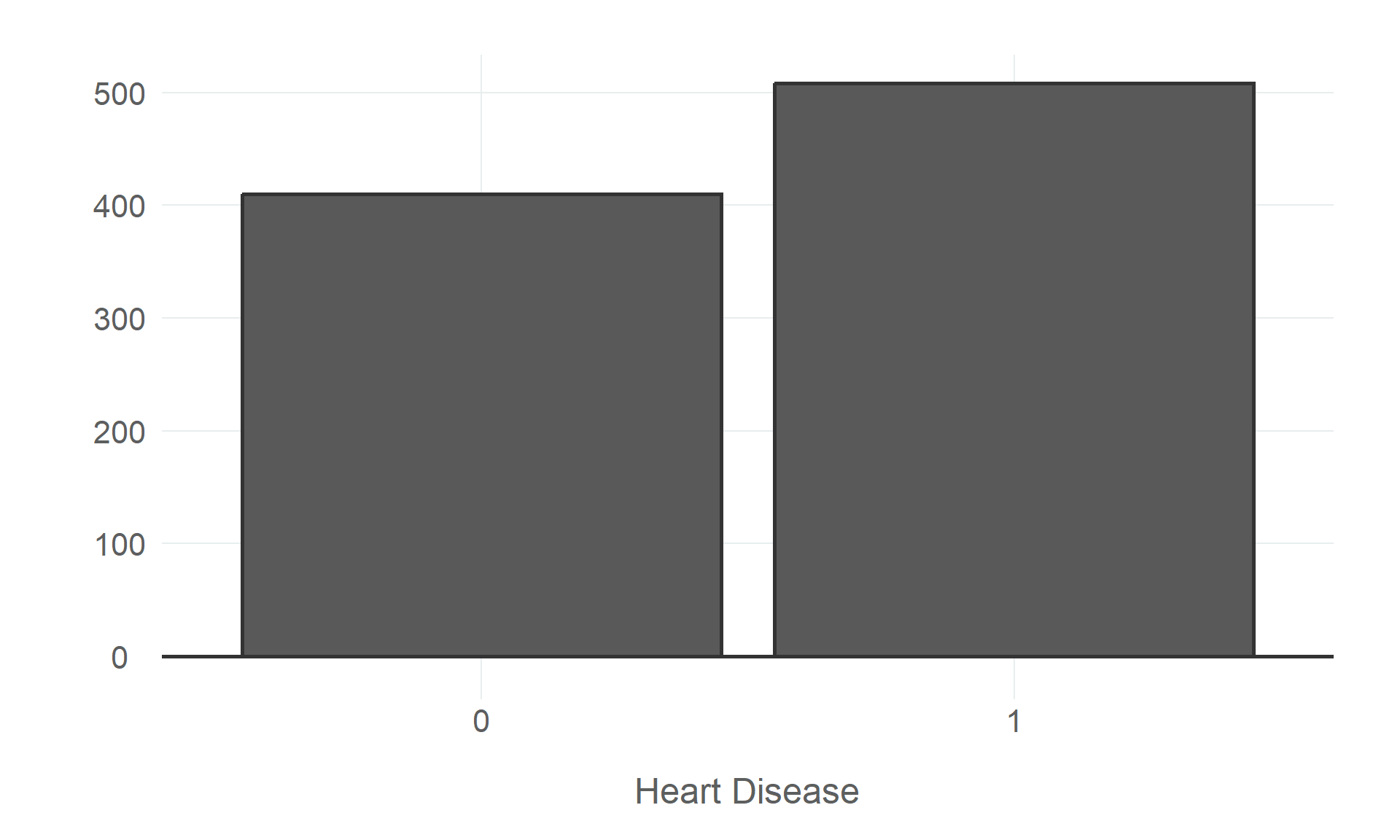
$ heart_disease <fct> 0, 1, 0, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 1, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1…